Sample Submission Guide
Sample Submission
- Complete and email the sample submission form and additional documents BEFORE shipping your samples to us.
- Ensure that your samples meet the quality and quantity requirement (see below). Samples which do not meet these criteria will not be processed.
- Prepare your samples in a 96-well plate or in a 1.5 ml tube format according to the guideline below. Failing to comply may lead to the delay in the processing and additional charge may apply.
- Please submit your samples TOGETHER with the sample submission form.
Sample Submission Form
Download Sample Submission Form
Sample Quality and Quantity Requirement
Once we receive your samples we will perform quality control analysis to determine the quality, concentration and volume of your samples to ensure that we have a sufficient amount. We strongly recommend that you perform your own sample QC to ensure the correct concentration of sample is shipped. This will help to expedite your project.
- At least 20 ul of 20 ng/ul of DNA is required. The concentration can be measured using nanodrop. If the concentration is lower than 20 ng/ul, please use Qubit or Picogreen for measuring the concentration.
- DNA must be double-stranded and not degraded as assessed by agarose gel electrophoresis (if there is sufficient DNA to perform this)
- Optical density ratio 260/280 of 1.8-2.0
- Optical density ratio 260/230 of 1.8-2.0
- Ensure the gDNA extracted is resuspended in nuclease-free water
Labelling and Handling of Samples
All samples should be securely sealed, including wrapping with parafilm membrane.
For small submission numbers (1-16 SAMPLES)
• Clearly label all sample tubes. • In order to avoid crushing during transfer which may cause sample loss, it is recommended to put the Eppendorf tube in a 50ml centrifuge tube or a similar container. Secure the tube with additional wrapping like cotton and absorbent paper.
For large submission numbers (17+ SAMPLES)
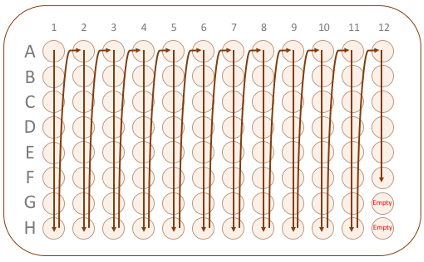
• Samples must be filled in the 96 well plates, and must be by columns, not rows. See the diagram below. Please leave the last two spots G12 and H12 empty for controls. • The plate must be clearly labelled on the skirt. • For shipping, protect the plates by surrounding the plates with strong rigid cardboard.
Transport
DNA samples can be shipped in room temperature. For RNA or tissue samples, please ship on dry ice. Depending upon the quality of the insulated shipping container and the samples, 3 to 5 kilograms of dry ice should be used for each 24-hour period. We recommend using a minimum of 3 kilograms and ideally 5 kilograms of dry ice. In addition, please try to ensure that the package is tightly sealed and labeled correctly.
By post:
Deepa Panicker
Sequencing Facility, AIMI
UTS Science Store
Level 2 Building 1
Thomas Street, Ultimo
NSW, Australia 2007
In Person:
Level 7 Building 4
Cnr of Thomas Street and Harris Street
Ultimo NSW
Drop off hour: Monday – Friday, 10 am – 3 pm
Sample Metadata Sheet
For data analysis, the metadata must be completed before any sequencing work on the samples can begin. Please fill in the excel spreadsheet (we will email it to you on request), and email it to the appropriate contact as soon as possible. Please follow the instruction within the spreadsheet, and if you are missing information for some of these fields please contact us for further advise.
Sample Quality Control and Re-submission
Once we receive your samples we will perform quality control analysis to determine the quality, concentration and volume of your samples to ensure that we have a sufficient amount. We strongly recommend that you perform your own sample QC to ensure the correct concentration of sample is shipped. This will help to expedite your project. If a sample(s) fails initial QC or do not comply with our sample submission guidelines, the client can either:
- Resubmit samples - If samples are provided in plates the entire plate must be resubmitted. The facility will not cherry pick samples.
- Arrange for samples to be returned and then resubmit - The client will arrange courier collection and liaise with the facility on the scheduling of this collection. The facility is not responsible for any courier fees incurred for sample return.
- Withdraw their submission - The client must pay the facility the costs of any consumables or other materials that have been ordered by the facility on their behalf in order to perform the services outlined in the quote.
In the case resubmitting after failed QC, the client must pay the cost of QC the resubmitted samples.
Sample Processing, Analysis and Data Delivery
Sample Processing
Once the samples are accepted, the UTS Sequencing facility will move forward to processing your samples. All samples will be discarded 30 days after data delivery. If you would like to keep your samples, please let us know and arrange the samples to be picked up as soon as possible.
Analysis
The facility will assess the data output to determine whether it has passed or failed. We provide the sequencing summary report including the data output, fastq files, fasta and .gfa files for assemblies, NCBI Bioproject numbers and NCBI Biosample numbers for samples submitted onto NCBI databases. Should you need any other file(s) that is not part of the standard deliverables please contact us and we will do our best to accommodate your request.
Data Delivery
Data is shared with the client through a download link that is emailed to the client. All data will be deleted 30 days after delivery. We will then move forward to invoicing for your project.
For further information please contact:
Joyce To
Email: joyce.to@uts.edu.au